Publications
Group highlights
(For a full list of publications and patents see below or go to Google Scholar
O’Meara MJ, Leaver-Fay A, Tyka M, Stein A, Houlihan K, DiMaio F, Bradley P, Kortemme T, Baker D, Snoeyink J, Kuhlman B
J. Chem. Theory Comput. (2015)
Lyu J, Wang S, Balius TE, Singh I, Levit A, Moroz YS, O’Meara MJ, Che T, Algaa E, Tolmachev AA, Shoichet BK, Roth BL, Irwin JJ
Gordon DE*, Jang GM*, Bouhaddou M*, Xu J, Obernier K*, White K*, O’Meara MJ*, et al. UCSF QBI Coronavirus Research Group
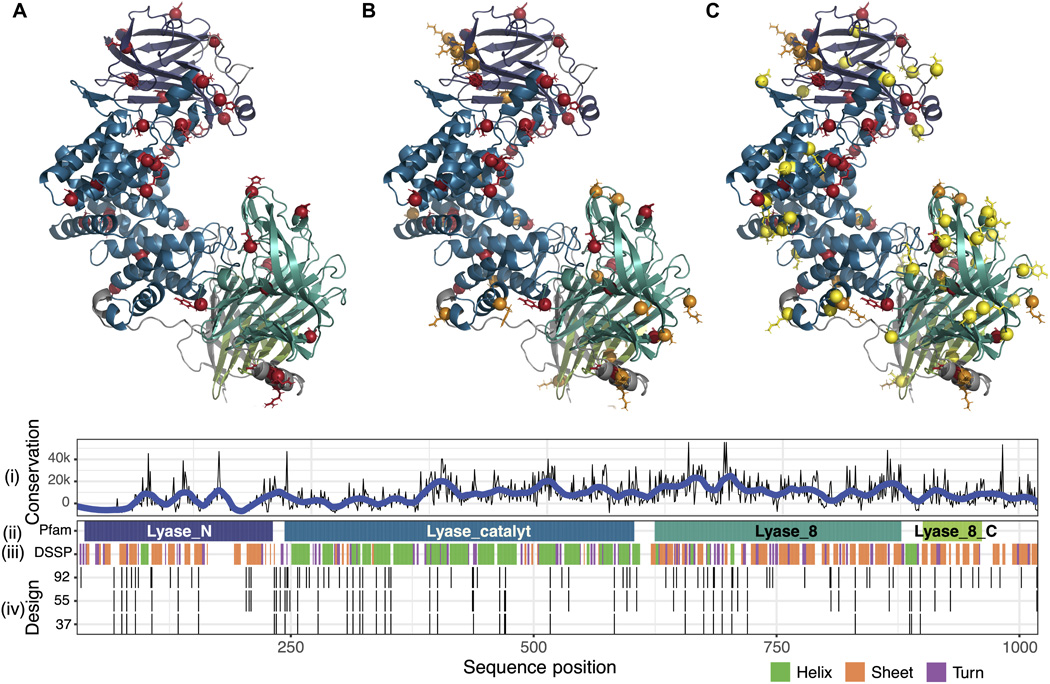
Hettiaratchi MH*, O’Meara MJ*, O’Meara TR, Pickering AJ, Letko-Khait N, Shoichet MS [* Contributed Equally]
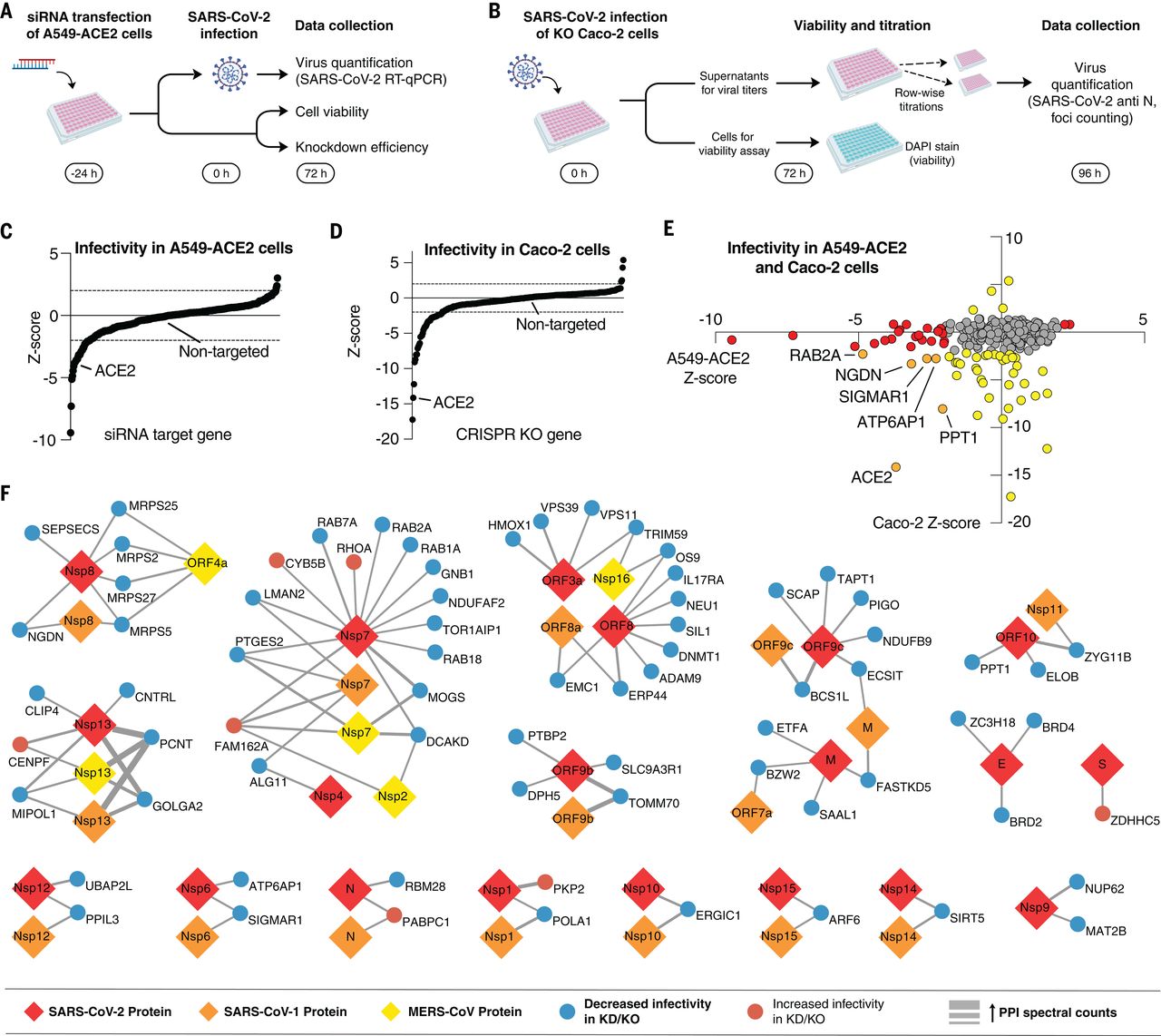
Gordon DE, et al., O'Meara MJ, et al., UCSF QBI Coronavirus Research Group
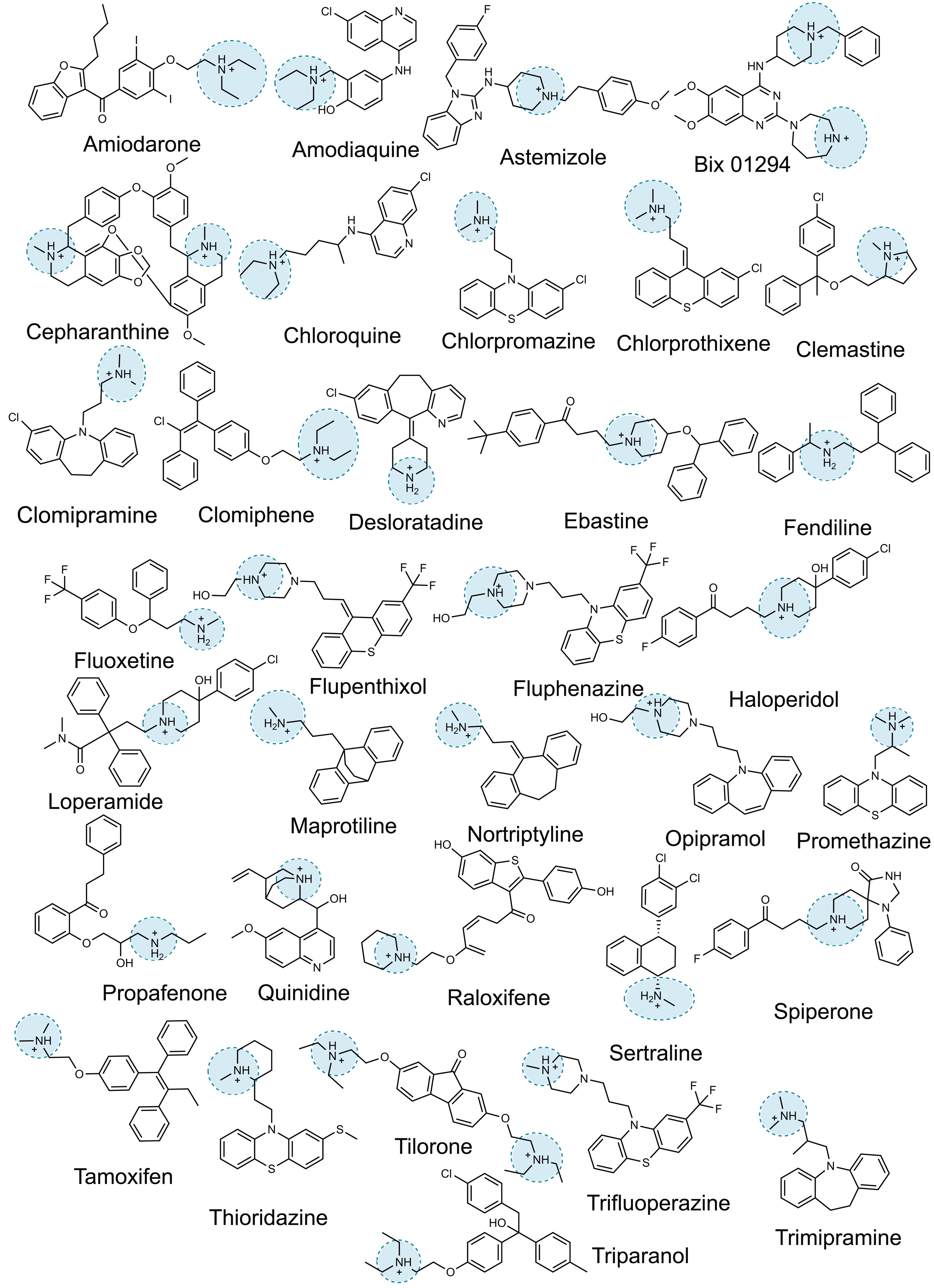
Tummino TA, Rezelj VV, Fischer B, Fischer A, O’Meara MJ, Monel B, Vallet T, Zhang Z, Alon A, O’Donnell HR, Lyu J, Schadt H, White KM, Krogan NJ, Urban L, Shokat KM, Kruse AC, García-Sastre A, Schwartz O, Moretti F, Vignuzzi M, Pognan F, Shoichet BK
Recommended in Faculty Opinions
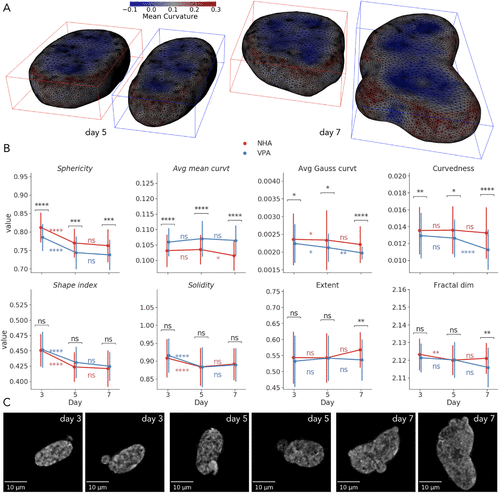
Kalinin AA, Hou X, Ade AS, Fon GV, Meixner W, Higgins GA, Sexton JZ, Xiang Wan, Dinov ID, O'Meara MJ, Athey BD
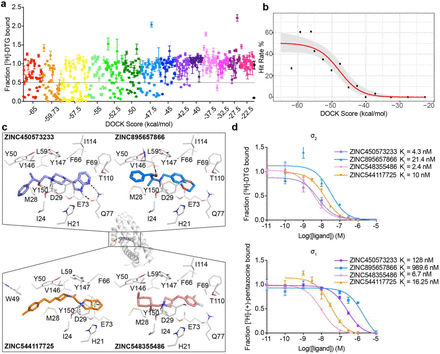
Alon A, Lyu J, Braz JM, Tummino TA, Craik V, O'Meara MJ, Webb CM, Radchenko DS, Moroz YS, Huang XP, Liu Y, Roth BL, Irwin JJ, Basbaum AI, Shoichet BK, Kruse AC
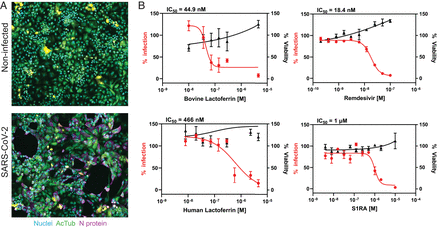
Mirabelli C, Wotring JW, Zhang CJ, McCarty SM, Fursmidt R, Pretto CD, Qiao Y, Zhang Y, Frum T, Kadambi NS, Amin AT, O’Meara TR, Spence JR, Huang J, Alysandratos KD, Kotton DN, Handelman SK, Wobus CE, Weatherwax KJ, Mashour GA, O’Meara MJ*, Chinnaiyan AM*, Sexton JZ* [*contributed equally]
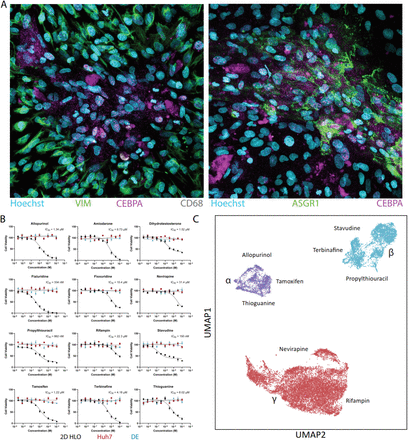
Zhang CJ, O’Meara MJ, Meyer SR, Huang S, Capeling MM, Ferrer-Torres D, Childs CJ, Spence JR, Fontana RJ, Sexton JZ.
Fu C, Zhang X, Veri AO, Lyer KR, Lash E, Xue A, Yan H, Revie NM, Wong C, Lin ZY, Polvi EJ, Liston SD, VanderSluis B, Hou J, Yashiroda Y, Gingras AC, Boone C, O'Meara TR, O'Meara MJ, Noble S, Robbins N, Myers CL, Cowen LE
Full List of publications
Colour Patterns for Polychromatic Four-colourings of Rectangular Subdivisions
Haverkort HJ, Löffler M, Mumford E, O’Meara MJ, Snoeyink J, Speckmann B
24th European Workshop on Computational Geometry (2008)
Maximum Geodesic Routing in the Plane with Obstacles
O’Meara MJ, Millman DL, Snoeyink J, Verma V
The Canadian Conference on Computational Geometry (2010)
Scientific Benchmarks for Guiding Macromolecular Force Field Improvement
Leaver-Fay A, O’Meara MJ, Tyka M, Jacak R, Song Y, Kellogg EH, Thompson J, Davis IW, Pache, RA, Lyskov S, Gray JJ, Kortemme T, Richardson JS, Havranek JJ, Snoeyink J, Baker D, Kuhlman B [*contributed equally]
Meth. Enzymol. (2013)
Role of Electrostatic Repulsion in Controlling pH-dependent Conformational Changes of Viral Fusion Proteins
Harrison JS, Higgins CD, O’Meara MJ, Koellhoffer JF, Kuhlman BA, & Lai JR
Structure (2013)
The Cryptococcus neoformans Rim101 Transcription Factor Directly Regulates Genes Required for Adaptation to the Host
O’Meara TR, Xu W, Selvig KM, O’Meara MJ, Mitchell AP, Alspaugh JA
Mol. Cell. Biol. (2014)
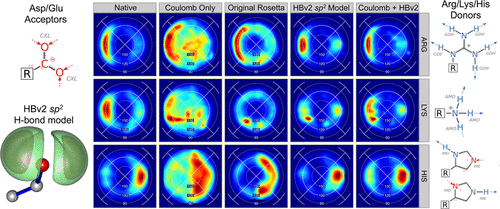
Combined Covalent-Electrostatic Model of Hydrogen Bonding Improves Structure Prediction with Rosetta
O’Meara MJ, Leaver-Fay A, Tyka M, Stein A, Houlihan K, DiMaio F, Bradley P, Kortemme T, Baker D, Snoeyink J, Kuhlman B
J. Chem. Theory Comput. (2015)
A Web Resource for Standardized Benchmark Datasets, Metrics, and Rosetta Protocols for Macromolecular Modeling and Design
Ó Conchúir S, Barlow KA, Pache RA, Ollikainen N, Kundert K, O’Meara MJ, Smith CA, Kortemme T
PLoS ONE (2015)
The recognition of identical ligands by unrelated proteins
Barelier S, Sterling T, O’Meara MJ, Shoichet BK
ACS Chem. Biol. (2015)
Ligand Similarity Complements Sequence, Physical Interaction, and Co-Expression for Gene Function Prediction
O’Meara MJ, Ballouz S, Shoichet BK, Gillis J
PLoS ONE (2016)
The Rosetta all-atom energy function for macromolecular modeling and design
Alford RF, Leaver-Fay A, Jeliazkov JR, O’Meara MJ, DiMaio FP, Park H, Shapovalov MV, Renfrew PD, Mulligan VK, Kappel K, Labonte JW, Pacella MS, Bonneau R, Bradley P, Dunbrack RL, Das R, Baker D, Kuhlman B, Kortemme T, Gray JJ
J. Chem. Theory Comput. (2017)
Integrative mapping of enzymatic pathways
Calhoun S, Korczynska M, Wichelecki DJ, San Francisco B, Zhao S, Rodionov DA, Vetting MW, Al-Obaidi NF, Lin H, O’Meara MJ, Scott DA, Morris JH, Russel D, Ferrin TE, Almo SC, Osterman AL, Jacobson MP, Gerlt JA, Shoichet BK, Sali A
eLife (2018)
High-throughput screening identifies genes required for fungal induction of macrophage pyroptosis
O’Meara TR, Duah K, Guo CX, Maxson ME, Gaudet R, Koselny K, Wellington M, Powers ME, MacAlpine J, O’Meara MJ, Veri AO, Grinstein S, Noble SM, Krysan D, Gray-Owen S, Cowen LE
mBio (2018)
Local Delivery of Stabilized Chondroitinase ABC Degrades Chondroitin Sulfate Proteoglycans in Stroke-Injured Rat Brains
Hettiaratchi MH, O’Meara MJ, Teal CJ, Payne SL, Pickering AJ, Shoichet M
J. Control Release (2018)
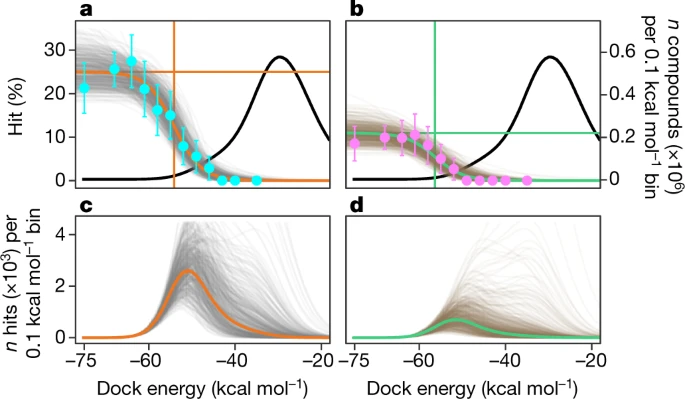
Ultra-large library docking for new chemotypes
Lyu J, Wang S, Balius TE, Singh I, Levit A, Moroz YS, O’Meara MJ, Che T, Algaa E, Tolmachev AA, Shoichet BK, Roth BL, Irwin JJ
Nature (2019)
Global Proteomic Analyses Define an Environmentally Contingent Hsp90 Interactome and Reveal Chaperone-Dependent Regulation of Stress Granule Proteins and the R2TP Complex in a Fungal Pathogen
O’Meara TR, O’Meara MJ, Polvi EJ, Pourhaghighi MR, Liston SD, Lin Z-Y, Veri AO, Emili A, Gingras A-C, Cowen LE
PLOS Biol. (2019)
A Unified Stochastic Gradient Approach to Designing Bayesian-Optimal Experiments
Foster A, Jankowiak M, O’Meara MJ, Teh YW, Rainforth T
AISTATS (2020)
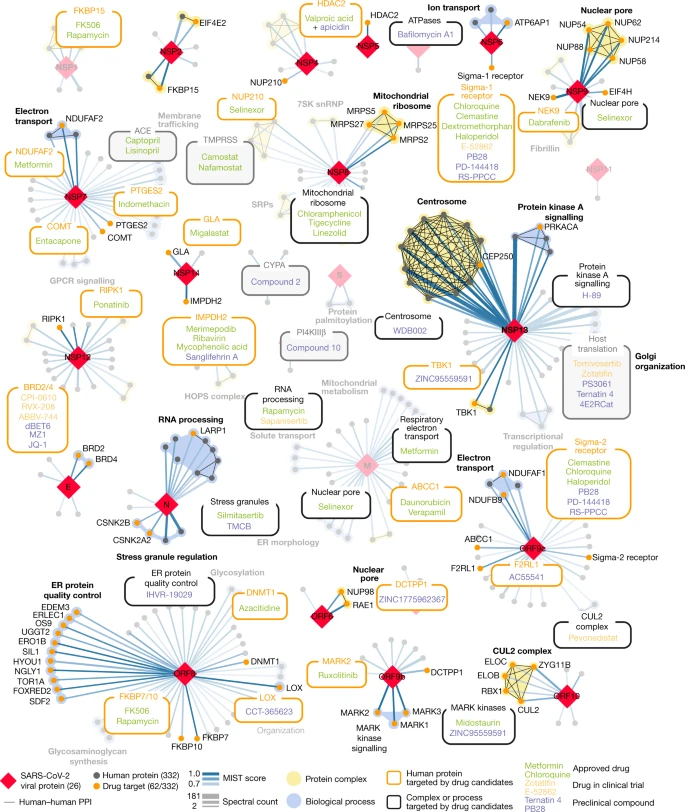
A SARS-CoV-2 protein interaction map reveals targets for drug repurposing
Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White K, O’Meara MJ, et al. UCSF QBI Coronavirus Research Group
Nature (2020)
Reengineering biocatalysts: Computational redesign of chondroitinase ABC improves efficacy and stability
Hettiaratchi MH, O’Meara MJ, O’Meara TR, Pickering AJ, Letko-Khait N, Shoichet MS [* Contributed Equally]
Sci. Adv. (2020)
Machine Learning and Assay Development for Image-based Phenotypic Profiling of Drug Treatments
Sexton JZ, Fursmidt R, O’Meara MJ, Rao A, Egan D, Haney SA
NCATS Assay Guidance Manual [Accepted] (2021)
Comparative Host-Coronavirus Protein Interaction Networks Reveal Pan-Viral Disease Mechanisms
Gordon DE, et al., O’Meara MJ, et al., UCSF QBI Coronavirus Research Group
Science (2020)
DeORFanizing Candida albicans Genes using Coexpression
O’Meara TR, O’Meara MJ
mSphere (2020)
Property-Unmatched Decoys in Docking Benchmarks
Stein RM, Yang Y, Balius ME, O’Meara MJ, Lyu J, Young J, Tang K, Shoichet BK, Irwin JJ
J. Chem. Inf. Model (2021)
Phospholipidosis is a shared mechanism underlying the in vitro antiviral activity of many repurposed drugs against SARS-CoV-2
Tummino TA, Rezelj VV, Fischer B, Fischer A, O’Meara MJ, Monel B, Vallet T, Zhang Z, Alon A, O’Donnell HR, Lyu J, Schadt H, White KM, Krogan NJ, Urban L, Shokat KM, Kruse AC, García-Sastre A, Schwartz O, Moretti F, Vignuzzi M, Pognan F, Shoichet BK
Science (2021)
Valproic Acid-Induced Changes of 4D Nuclear Morphology in Astrocyte Cells
Kalinin AA, Hou X, Ade AS, Fon GV, Meixner W, Higgins GA, Sexton JZ, Xiang Wan, Dinov ID, O’Meara MJ, Athey BD
Mol. Biol. Cell (2021)
Crystal structures of the σ2 receptor template large-library docking for selective chemotypes active in vivo
Alon A, Lyu J, Braz JM, Tummino TA, Craik V, O’Meara MJ, Webb CM, Radchenko DS, Moroz YS, Huang XP, Liu Y, Roth BL, Irwin JJ, Basbaum AI, Shoichet BK, Kruse AC
BioRxiv (2021)
Morphological cell profiling of SARS-CoV-2 infection identifies drug repurposing candidates for COVID-19
Mirabelli C, Wotring JW, Zhang CJ, McCarty SM, Fursmidt R, Pretto CD, Qiao Y, Zhang Y, Frum T, Kadambi NS, Amin AT, O’Meara TR, Spence JR, Huang J, Alysandratos KD, Kotton DN, Handelman SK, Wobus CE, Weatherwax KJ, Mashour GA, O’Meara MJ, Chinnaiyan AM, Sexton JZ* [*contributed equally]
PNAS (2021)
A Multi-Omics Human Liver Organoid Screening Platform for DILI Risk Prediction.
Zhang CJ, O’Meara MJ, Meyer SR, Huang S, Capeling MM, Ferrer-Torres D, Childs CJ, Spence JR, Fontana RJ, Sexton JZ.
BioRxiv (2021)
Chondroitinase ABC Mutants and Methods of Manufacture and use Thereof
Shoichet MS, Hettiaratchi MH, O’Meara TR, O’Meara MJ
WO/2021/159205
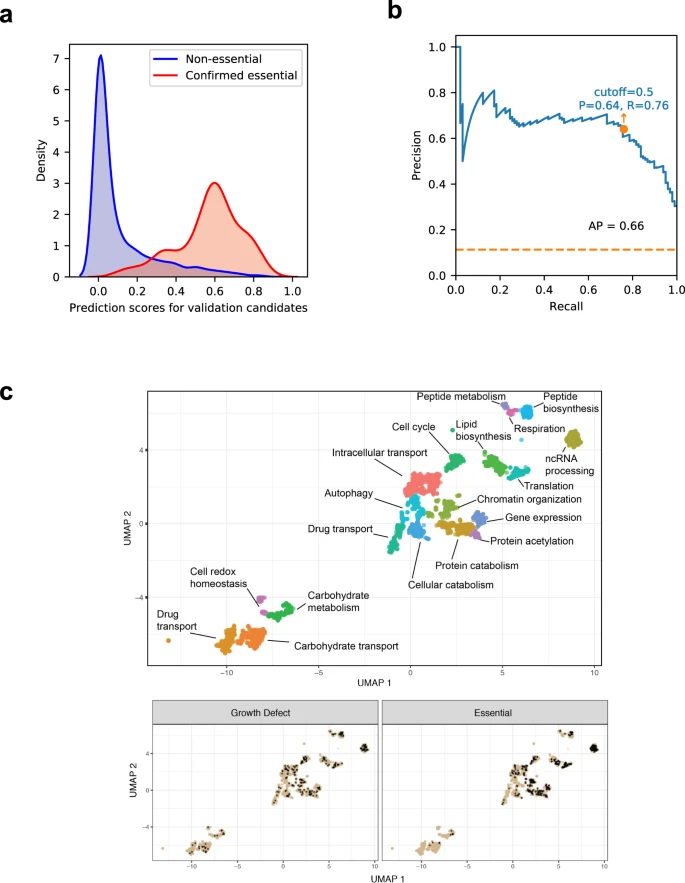
Leveraging machine learning essentiality predictions and chemogenomic interactions to identify antifungal targets
Fu C, Zhang X, Veri AO, Lyer KR, Lash E, Xue A, Yan H, Revie NM, Wong C, Lin ZY, Polvi EJ, Liston SD, VanderSluis B, Hou J, Yashiroda Y, Gingras AC, Boone C, O’Meara TR, O’Meara MJ, Noble S, Robbins N, Myers CL, Cowen LE
Nat. Comm (2021)
Structures of the σ2 receptor enable docking for bioactive ligand discovery
Alon A, Lyu J, Braz JM, Tummino TA, Craik V, O’Meara MJ, Webb CM, Radchenko DS, Moroz YS, Huang X, Liu Y, Roth BL, Irwin JJ, Basbaum AI, Shoichet BK, Kruse AC
Nature (2021)
Multiple ParA/MinD ATPases coordinate the positioning of disparate cargos in a bacterial cell
Pulianmackal LT, Limcaoco JMI, Ravi K, Yang S, Zhang J, Tran MK, O’Meara MJ, Vecchiarelli AG
Nat. Commun. (2022)
Development of an automated screen for Kv7.2 potassium channels and discovery of a new agonist chemotype
Hernandez CC, Tarfa RA, Limcaoco JMI, Liu R, Mondal P, Hill C, Duncan RK, Tzounopoulos T, Stephenson CR, O’Meara MJ, Wipf P
Bioorganic & Medicinal Chemistry Letters (2022)
In Vitro Evaluation and Mitigation of Niclosamide’s Liabilities as a COVID-19 Treatment
Wotring JW, McCarty SM, Shafiq K, Zhang CJ, Nguyen T, Meyer SR, Fursmidt R, Mirabelli C, Clasby MC, Wobus CE, O’Meara MJ, Sexton JZ
Vaccines (2022)
Prioritizing virtual screening with interpretable interaction fingerprints
Fassio AV, Shub L, Ponzoni L, McKinley J, O’Meara MJ, Ferreira RS, Keiser MJ, Minardi RCM
J. Chem. Inf. Model. (2022)
High-Content Screening to Identify Inhibitors of Dengue Virus Replication
Hoffstadt JG, Wotring JW, Porter S, Halligan BS, O’Meara MJ, Tai AW, Sexton JZ
bioArxiv (2023)
Predicting the Structural Impact of Human Alternative Splicing
Song Y, Zhang C, Omenn GS, O’Meara MJ, Welch JD
bioRxiv (2023)
A Human Liver Organoid Screening Platform for DILI Risk Prediction
Zhang CJ, Meyer SR, O’Meara MJ, Huang S, Capeling MM, Ferrer-Torres D, Childs CJ, Spence JR, Fontana RJ, Sexton JZ
J. Hepatol. (2023)
The life and times of endogenous opioid peptides: Updated understanding of synthesis, spatiotemporal dynamics, and the clinical impact in alcohol use disorder
Margolis EB, Moulton MG, Lambeth PS, O’Meara MJ
Neuropharmacology (2023)
Machine Learning and Assay Development for Image-based Phenotypic Profiling of Drug Treatments
Sexton JZ, Fursmidt R, O’Meara MJ, Omta W, Rao A, Egan DA, Haney SA
Eli Lilly & Company and the National Center for Advancing Translational Sciences (2023)
Candida albicans selection for human commensalism results in substantial within-host diversity without decreasing fitness for invasive disease
Anderson FM, Visser ND, Amses KR, Hodgins-Davis A, Weber AM, Metzner KM, McFadden MJ, Mills RE, O’Meara MJ, James TY, O’Meara TR
PLoS Biology (2023)
Imaging-Based Screening Identifies Modulators of the eIF3 Translation Initiation Factor Complex in Candida albicans
Metzner K, O’Meara MJ, Halligan B, Wotring JW, Sexton JZ, O’Meara TR
Antimicrob. Agents Chemother. (2023)
LUMIC: Latent diffUsion for Multiplexed Images of Cells
Hung AZ, Zhang CJ, Sexton JZ, O’Meara MJ, Welch JD
bioRxiv (2024)
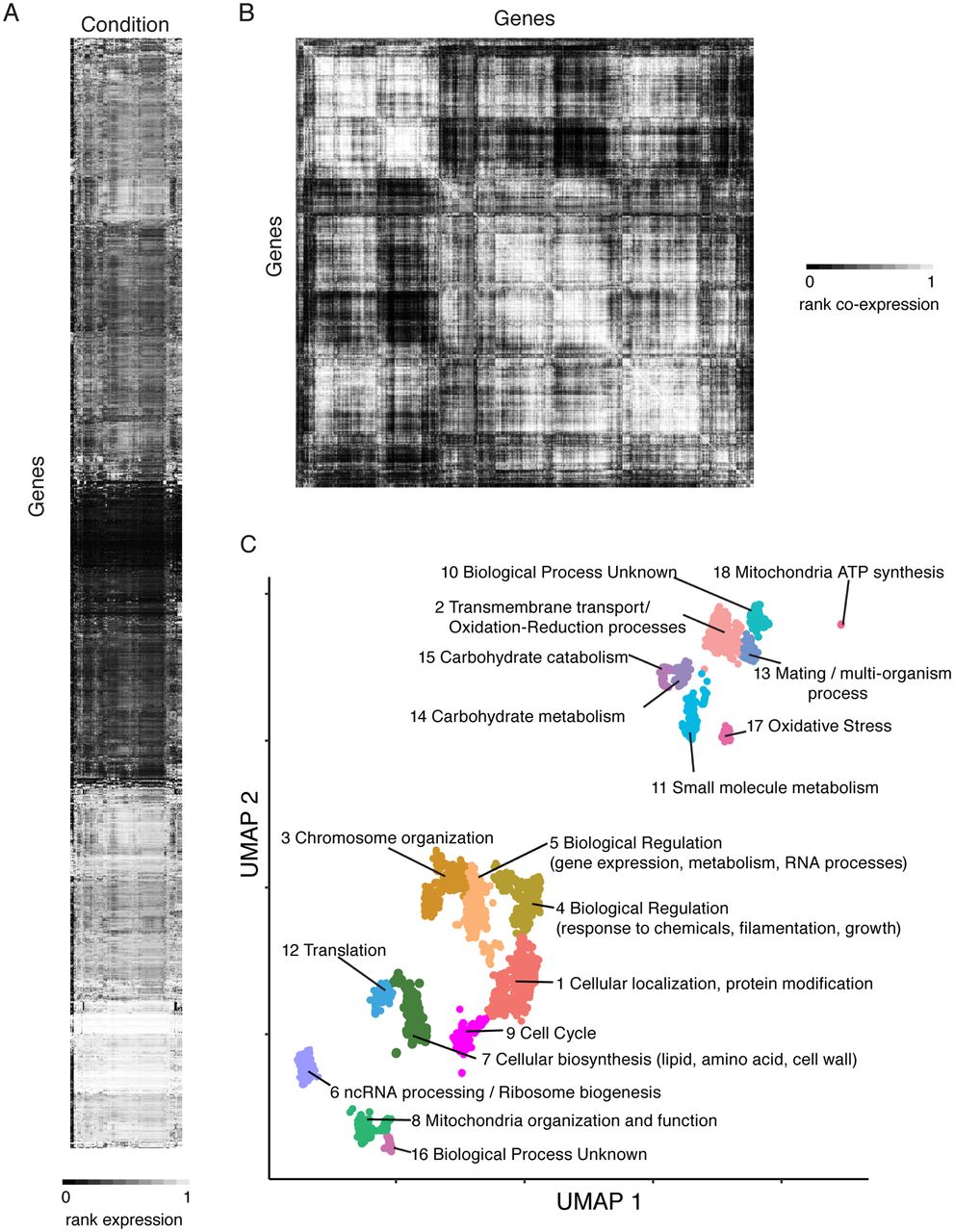
CryptoCEN: A Co-Expression Network for Cryptococcus neoformans reveals novel proteins involved in DNA damage repair
O’Meara MJ, Rapala JR, Nichols CB, Alexandre AC, Billmyre RB, Steenwyk JL, Alspaugh JA, O’Meara TR
Plos Genetics (2024)